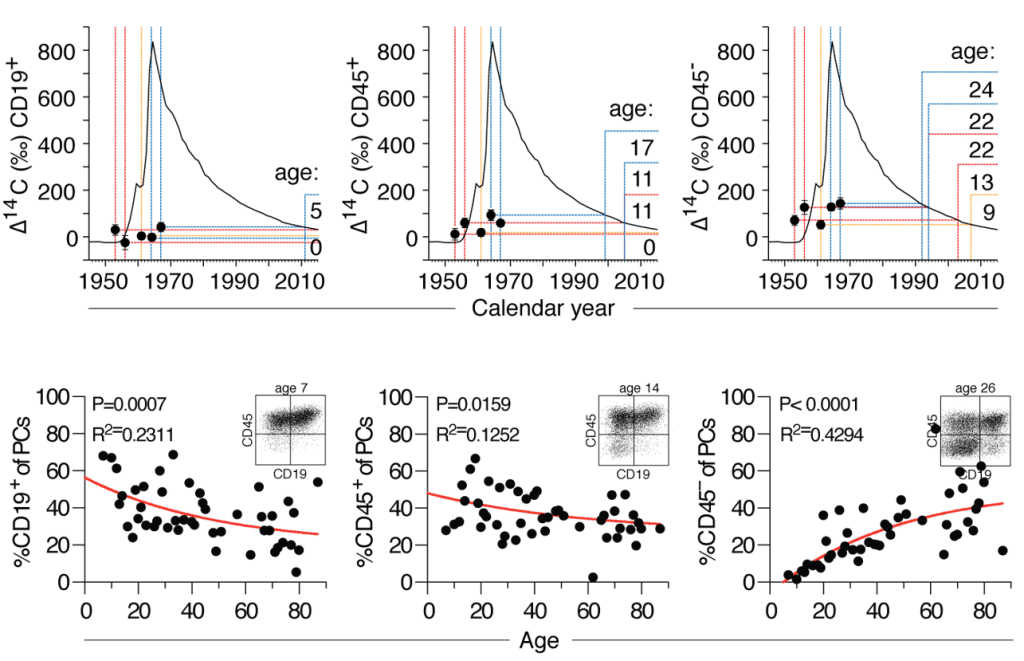
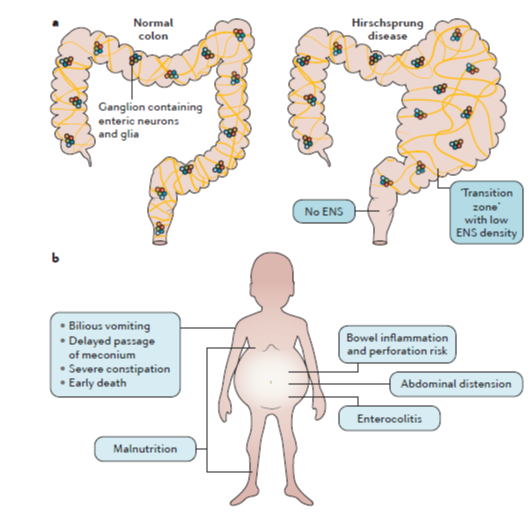
We seek a motivated and enthusiastic co-worker for a 3-year PhD fellowship (with a possibility of extension) in our research group (https://jahnsenlab.org/).
The position will primarily be based at the Department of Pathology, Oslo University Hospital/Rikshospitalet.
To read more about position click here.